The scope of health data generated is dramatically growing in societal importance and demand for it will continue to rise. Collaborative new research is required, in both the access and application methods, that allows for computing upon these data sets in context, and in secure and responsive ways.
Future use of these data will require new technical architectures that support the legal requirements and the range of evolving users. CeDAR participates in the field of health data research through a focus on two components: privacy-protecting machine-learning and synthetic data generation and portability.
Endovascular Perfusion Augmentation for Critical Care (EPACC): Personalized and Adaptive Therapy for Resuscitation After Trauma
Chen-Neeh Chuah
Post-injury resuscitation is predicated on maintaining adequate systemic perfusion via balanced blood transfusion, crystalloids, and vasopressors. Yet, these medical supplies are often limited in resource-constrained environments. Furthermore, the care of critically injured patients during damage control resuscitation requires continuous patient management at the bedside to optimize outcomes. This project aims to develop a novel resuscitation platform that optimizes critical care management in real time. By harnessing the power of endovascular devices with AI assisted fluid and medication delivery, this platform can provide highly nuanced, dynamic and patient-centered precision critical care beyond what is possible by conventional means. This is a multi-institution project with collaborators from Wake Forest School of Medicine, the United States Air Force, and the Naval Medical Research.
Intelligent Learning based Critical Care (ICCare): Real-time Analytic and Intelligent Learning Platform for Diagnostic Test and Patient Management in Critical Care
Chen-Neeh Chuah
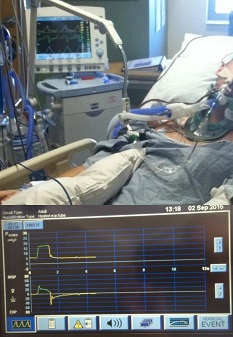 Hospital Intensive Care Units (ICUs) has been known as a data-rich, information-poor environment. Multitude of sensors and monitors are often present to monitor the patients' conditions, but the data collected are either not stored and analyzed continuously, or exist in isolated clusters that are not well-integrated. In addition, electronic medical record (EMR) systems often contain unstructured, subjective data from patient histories (e.g., clinician notes, X-ray reports) with limited automation, timeliness, and reproducibility. A real-time analytic platform is essential to integrate these heterogeneous sources of data in a meaningful way that can be fed into intelligent learning algorithms to derive predictive models that can be useful for automating diagnostic tests, detection of patients' states, or aid in making clinical decisions.
Hospital Intensive Care Units (ICUs) has been known as a data-rich, information-poor environment. Multitude of sensors and monitors are often present to monitor the patients' conditions, but the data collected are either not stored and analyzed continuously, or exist in isolated clusters that are not well-integrated. In addition, electronic medical record (EMR) systems often contain unstructured, subjective data from patient histories (e.g., clinician notes, X-ray reports) with limited automation, timeliness, and reproducibility. A real-time analytic platform is essential to integrate these heterogeneous sources of data in a meaningful way that can be fed into intelligent learning algorithms to derive predictive models that can be useful for automating diagnostic tests, detection of patients' states, or aid in making clinical decisions.
In our pilot project, we focus on developing an AI-driven diagnostic and prognosis tests for Acute-Respiratory Distress Syndrome (ARDS) Patients requiring mechanical ventilator machines in Critical Care. While mechanical ventilation (MV) machine is a life-saving therapy for patients in intensive care units (ICUs), it has been conclusively demonstrated in clinical studies that inappropriate delivery of MV can directly injure the lungs, leading to a condition called Acute-Respiratory Distress Syndrome (ARDS) and higher mortality. Attempts to automate ARDS diagnosis using rule-based computer algorithms have seen only limited success, requiring complex informatics infrastructures unavailable in current electronic medical record systems. We propose to employ advanced statistical and machine learning techniques to build an AI model that will operate on objective, readily available, high-dimensionality data including continuous ventilator waveform signals, electronic medical records (EMR), and wearable sensor data, to improve early recognition of ARDS. Early identification of ARDS patients, when they are most likely to benefit from ARDS-specific treatments, will lead to better overall outcomes that can be quantified (e.g., shorter hospital stay, lower mortality rate, mobility index). We expect that the research will lead to the refinement and validation of a functional automated ARDS diagnostic test ready for prospective evaluation in a large, multicenter cohort study aimed at improving the accuracy of ARDS diagnosis.
ML-VIRSA: Machine-Learning based Video Screening for Detecting Autism Risk in the First Year of Life
Chen-Neeh Chuah
Over the last two decades, the average age of first diagnosis of autism spectrum disorder (ASD) in the United States has remained steady, at over 4 years of age, despite an average age of first parental concerns of 14 months and recent progress in understanding manifestations of ASD in infancy. Early intensive intervention has been shown to be highly promising for young children with ASD, including infants and toddlers, but is typically reserved for children with a formal diagnosis, making accurate identification as early as possible imperative. A measure that could identify ASD risk during this period of onset offers the opportunity for intervention before the full set of symptoms is present.
PI Ozonoff and her team have developed a new video-based screening tool, the Video-referenced Infant Rating System for Autism (VIRSA), that utilizes a large library of video clips depicting a wide range of social-communication ability and relies solely on video in the ratings, with no written descriptions of behavior. We hypothesized that the semantic clarity afforded by video would provide improved early discrimination of infants at highest risk for ASD. The VIRSA was able to predict ASD diagnosis with high sensitivity across ages (6-18 months) and demonstrated 100% sensitivity (no false negatives) in concurrently identifying children showing signs of autism at 18 months of age.
Despite the demonstrated success of video-based screening, one major obstacle is the labor-intensive process of labeling and reviewing the videos manually by clinicians. In this proposed collaborative project, we propose to leverage videos from the VIRSA as a training set and apply computer vision and machine learning methods to develop a neural network model for ASD recognition.
Transabdominal Fetal Oximetry
Soheil Ghiasi
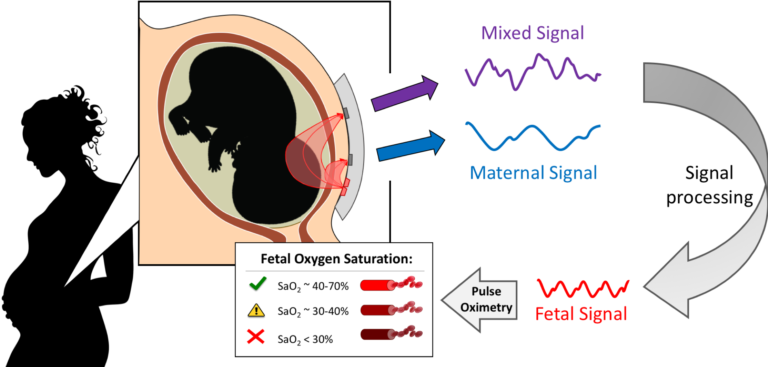
We are developing a non-invasive, transabdominal fetal oximeter, to provide worried mothers and obstetricians needed information regarding fetal well-being, namely fetal oxygenation, to reduce the number of unnecessary emergency C-sections. This is done by transmitting light in the near-infrared spectrum through the maternal abdomen and fetus to measure the oxygen saturation in the fetal blood using pulse oximetry. Pulse oximetry is a technique that utilizes the light absorption spectra of oxy- and deoxy- hemoglobin and captures a photoplethysmograph (PPG) from the vascular pulsations of the cardiac cycle. This PPG signal can then be used to calculate the oxygen saturation of the blood based on how much light was detected per heartbeat. In our case, a mixed maternal-fetal PPG is captured since light is traversing both maternal and fetal tissue, to which the fetal contribution can be isolated through Fourier analysis and filtering.

